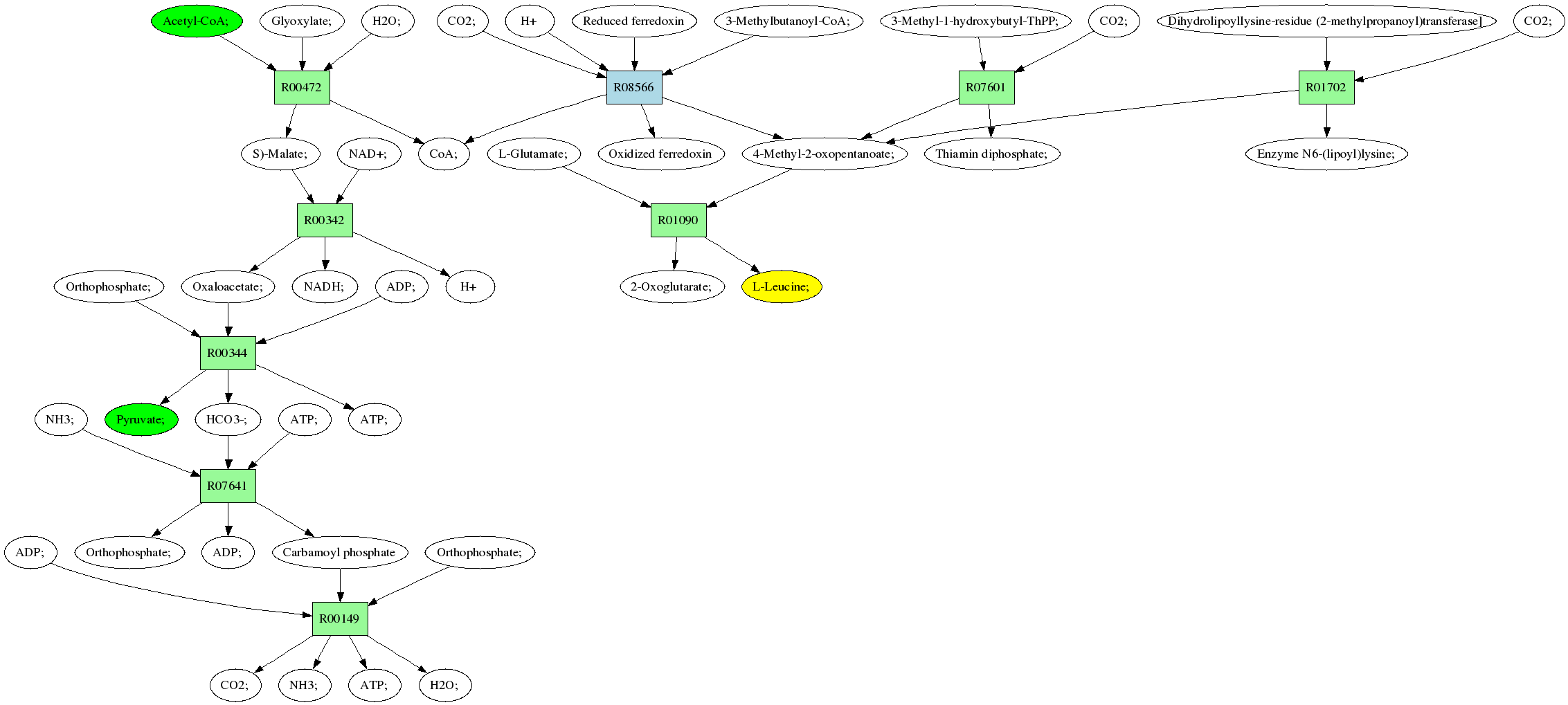
Back to all pathways
Sources: Pyruvate; (C00022)
Acetyl-CoA; (C00024)
Target:L-Leucine; (C00123)
Z: 0.166666666667
Composite map: C00024->C00123:[49->3]
Showing 1 best reactions for each rpair.
| RPAIR | Reaction | Score | Seq1 | Seq2 | Evalue | ECs | Equation |
|---|---|---|---|---|---|---|---|
| RP06023 | R00149 | 1431 | TRIRE0055928 | P31327|CPSM_HUMAN | 0.0 | 6.3.4.16 | CO2; + NH3; + ATP; + H2O; <=> Carbamoyl phosphate + Orthophosphate; + ADP; |
| RP05859 | R01702 | 414 | TRIRE0056726 | Q8HXY4|ODBA_MACFA | e-115 | 1.2.4.4 | 4-Methyl-2-oxopentanoate; + Enzyme N6-(lipoyl)lysine; <=> CO2; + Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase] |
| RP06172 | R00472 | 966 | TRIRE0079271 | P28345|MASY_NEUCR | 0.0 | 2.3.3.9 | CoA; + S)-Malate; <=> Acetyl-CoA; + Glyoxylate; + H2O; |
| RP05067 | R00344 | 1827 | TRIRE0021957 | O93918|PYC_ASPTE | 0.0 | 6.4.1.1 | HCO3-; + ATP; + Pyruvate; <=> Oxaloacetate; + Orthophosphate; + ADP; |
| RP00424 | R01090 | 480 | TRIRE0045459 | P47176|BCA2_YEAST | e-135 | 2.6.1.42 | 2-Oxoglutarate; + L-Leucine; <=> L-Glutamate; + 4-Methyl-2-oxopentanoate; |
| RP11035 | R01702 | 414 | TRIRE0056726 | Q8HXY4|ODBA_MACFA | e-115 | 1.2.4.4 | 4-Methyl-2-oxopentanoate; + Enzyme N6-(lipoyl)lysine; <=> CO2; + Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase] |
| RP10992 | R07641 | 3256 | TRIRE0055928 | O93937|PYR1_EMENI | 0.0 | 6.3.5.5,2.1.3.2 | NH3; + ATP; + HCO3-; <=> Carbamoyl phosphate + Orthophosphate; + ADP; |
| RP01666 | R08566 | 41 | TRIRE0081576 | O58413|VORA_PYRHO | 0.011 | 1.2.7.7 | CoA; + 4-Methyl-2-oxopentanoate; + Oxidized ferredoxin <=> 3-Methylbutanoyl-CoA; + CO2; + H+ + Reduced ferredoxin |
| RP00079 | R00342 | 427 | TRIRE0123729 | P17505|MDHM_YEAST | e-119 | 1.1.1.37 | S)-Malate; + NAD+; <=> Oxaloacetate; + NADH; + H |
| RP11034 | R07601 | 414 | TRIRE0056726 | Q8HXY4|ODBA_MACFA | e-115 | 1.2.4.4 | Thiamin diphosphate; + 4-Methyl-2-oxopentanoate; <=> CO2; + 3-Methyl-1-hydroxybutyl-ThPP; |
Pathway diagram (see pathway with molecule structures instead)