Back to all pathways
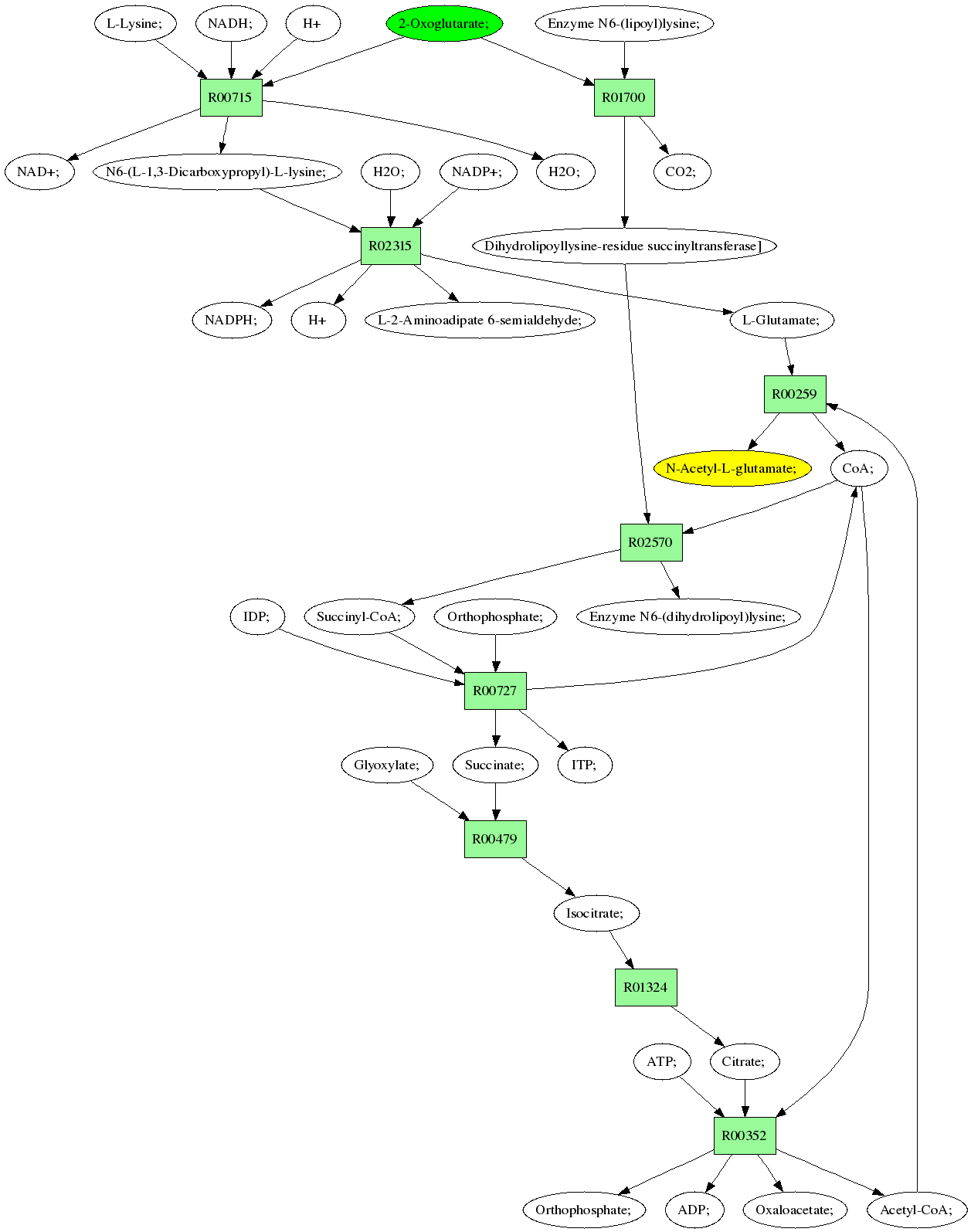
Sources: 2-Oxoglutarate; (C00026)
Target:N-Acetyl-L-glutamate; (C00624)
Z: 1.0
Composite map: C00026->C00624:[1->1,2->2,3->4,5->10,5->5,8->6,8->9]
Showing 1 best reactions for each rpair.
| RPAIR | Reaction | Score | Seq1 | Seq2 | Evalue | ECs | Equation |
|---|---|---|---|---|---|---|---|
| RP00602 | R02315 | 707 | TRIRE0123631 | Q9P4R4|LYS9_MAGGR | 0.0 | 1.5.1.10 | NADP+; + H2O; + N6-(L-1,3-Dicarboxypropyl)-L-lysine; <=> L-Glutamate; + L-2-Aminoadipate 6-semialdehyde; + NADPH; + H |
| RP00032 | R00727 | 696 | TRIRE0080881 | Q9P567|SUCB_NEUCR | 0.0 | 6.2.1.4 | CoA; + Succinate; + ITP; <=> Succinyl-CoA; + IDP; + Orthophosphate; |
| RP04458 | R00259 | 380 | TRIRE0077242 | Q04728|ARGJ_YEAST | e-105 | 2.3.1.35,2.3.1.1 | L-Glutamate; + Acetyl-CoA; <=> CoA; + N-Acetyl-L-glutamate; |
| RP05880 | R00352 | 1107 | TRIRE0121824 | Q8X097|ACL1_NEUCR | 0.0 | 2.3.3.8 | CoA; + Citrate; + ATP; <=> Oxaloacetate; + Acetyl-CoA; + Orthophosphate; + ADP; |
| RP00930 | R00479 | 926 | TRIRE0021758 | Q8J232|ACEA_MAGGR | 0.0 | 4.1.3.1 | Isocitrate; <=> Succinate; + Glyoxylate; |
| RP05855 | R00715 | 366 | TRIRE0119788 | Q7SFX6|LYS1_NEUCR | e-101 | 1.5.1.7 | NAD+; + H2O; + N6-(L-1,3-Dicarboxypropyl)-L-lysine; <=> H+ + 2-Oxoglutarate; + L-Lysine; + NADH; |
| RP10932 | R02570 | 344 | TRIRE0003653 | P19262|ODO2_YEAST | 1e-94 | 2.3.1.61 | Succinyl-CoA; + Enzyme N6-(dihydrolipoyl)lysine; <=> CoA; + Dihydrolipoyllysine-residue succinyltransferase] |
| RP08475 | R00259 | 380 | TRIRE0077242 | Q04728|ARGJ_YEAST | e-105 | 2.3.1.35,2.3.1.1 | L-Glutamate; + Acetyl-CoA; <=> CoA; + N-Acetyl-L-glutamate; |
| RP01431 | R01324 | 1095 | TRIRE0077336 | P19414|ACON_YEAST | 0.0 | 4.2.1.3 | Citrate; <=> Isocitrate; |
| RP10771 | R01700 | 1293 | TRIRE0050531 | P20967|ODO1_YEAST | 0.0 | 1.2.4.2 | 2-Oxoglutarate; + Enzyme N6-(lipoyl)lysine; <=> CO2; + Dihydrolipoyllysine-residue succinyltransferase] |
Pathway diagram (see pathway with molecule structures instead)