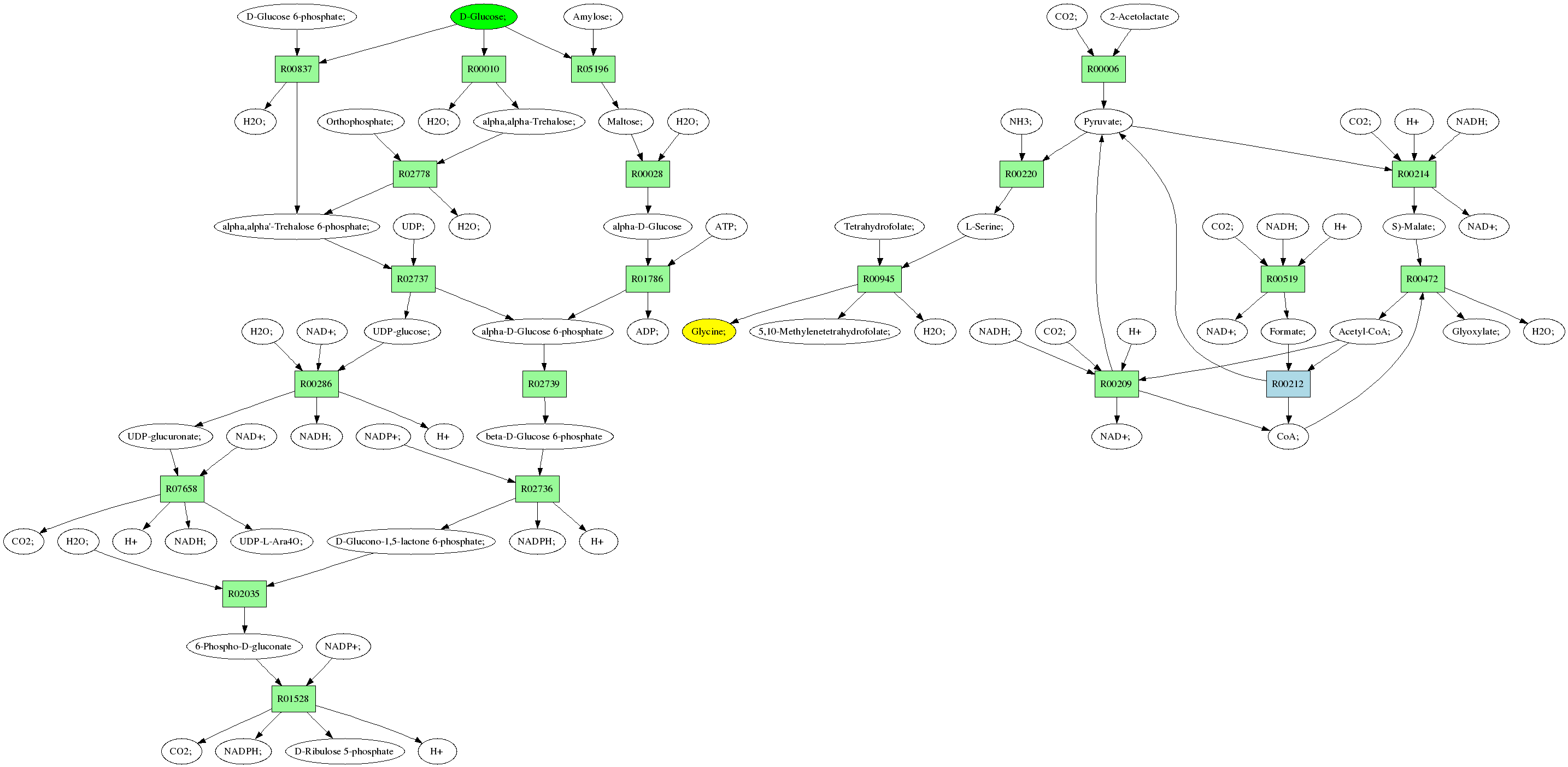
Back to all pathways
Sources: D-Glucose; (C00031)
Target:Glycine; (C00037)
Z: 1.0
Composite map: C00031->C00037:[4->1,4->2]
Showing 1 best reactions for each rpair.
| RPAIR | Reaction | Score | Seq1 | Seq2 | Evalue | ECs | Equation |
|---|---|---|---|---|---|---|---|
| RP06552 | R07658 | 1397 | TRIRE0002392 | Q01373|FOX2_NEUCR | 0.0 | 4.2.1.-,1.1.1.- | UDP-glucuronate; + NAD+; <=> CO2; + H+ + NADH; + UDP-L-Ara4O; |
| RP01134 | R00837 | 487 | TRIRE0108477 | P39795|TREC_BACSU | e-137 | 3.2.1.93 | alpha,alpha'-Trehalose 6-phosphate; + H2O; <=> D-Glucose 6-phosphate; + D-Glucose; |
| RP01925 | R02035 | 251 | TRIRE0073903 | O74455|6PGL_SCHPO | 9e-67 | 3.1.1.31 | D-Glucono-1,5-lactone 6-phosphate; + H2O; <=> 6-Phospho-D-gluconate |
| RP05698 | R00006 | 793 | TRIRE0074123 | P07342|ILVB_YEAST | 0.0 | 2.2.1.6 | CO2; + 2-Acetolactate <=> Pyruvate; |
| RP00453 | R00010 | 1325 | TRIRE0123456 | P78617|TREA_EMENI | 0.0 | 3.2.1.28 | alpha,alpha-Trehalose; + H2O; <=> D-Glucose; |
| RP06101 | R00214 | 527 | TRIRE0062153 | P36013|MAOM_YEAST | e-149 | 1.1.1.38 | S)-Malate; + NAD+; <=> CO2; + H+ + NADH; + Pyruvate; |
| RP02961 | R00028 | 595 | TRIRE0121351 | Q8TET4|GANC_HUMAN | e-169 | 3.2.1.20 | Maltose; + H2O; <=> alpha-D-Glucose |
| RP00285 | R00209 | 576 | TRIRE0077373 | O00087|DLDH_SCHPO | e-164 | 1.8.1.4 | CoA; + NAD+; + Pyruvate; <=> Acetyl-CoA; + H+ + NADH; + CO2; |
| RP05915 | R02737 | 729 | TRIRE0077602 | O59921|TPS1_EMENI | 0.0 | 2.4.1.15 | UDP-glucose; + alpha-D-Glucose 6-phosphate <=> alpha,alpha'-Trehalose 6-phosphate; + UDP; |
| RP00301 | R00945 | 819 | TRIRE0121686 | P34898|GLYC_NEUCR | 0.0 | 2.1.2.1 | 5,10-Methylenetetrahydrofolate; + H2O; + Glycine; <=> L-Serine; + Tetrahydrofolate; |
| RP00343 | R02778 | 677 | TRIRE0048707 | P31688|TPS2_YEAST | 0.0 | 3.1.3.12 | alpha,alpha'-Trehalose 6-phosphate; + H2O; <=> alpha,alpha-Trehalose; + Orthophosphate; |
| RP06172 | R00472 | 966 | TRIRE0079271 | P28345|MASY_NEUCR | 0.0 | 2.3.3.9 | CoA; + S)-Malate; <=> Acetyl-CoA; + Glyoxylate; + H2O; |
| RP03735 | R00519 | 637 | TRIRE0121298 | Q07103|FDH_NEUCR | 0.0 | 1.2.1.2 | NAD+; + Formate; <=> CO2; + NADH; + H |
| RP01590 | R05196 | 1389 | TRIRE0057128 | Q06625|GDE_YEAST | 0.0 | 2.4.1.25,3.2.1.33 | D-Glucose; + Amylose; <=> Maltose; |
| RP04290 | R00220 | 548 | TRIRE0004064 | O42615|THDH_ARXAD | e-156 | 4.3.1.19 | L-Serine; <=> NH3; + Pyruvate; |
| RP04232 | R00212 | 38 | TRIRE0082516 | Q46266|PFL_CLOPA | 0.19 | 2.3.1.54 | Acetyl-CoA; + Formate; <=> CoA; + Pyruvate; |
| RP02450 | R02736 | 889 | TRIRE0075769 | P48826|G6PD_ASPNG | 0.0 | 1.1.1.49 | NADP+; + beta-D-Glucose 6-phosphate <=> NADPH; + D-Glucono-1,5-lactone 6-phosphate; + H |
| RP04543 | R00286 | 416 | TRIRE0123705 | Q96558|UGDH_SOYBN | e-116 | 1.1.1.22 | NAD+; + UDP-glucose; + H2O; <=> UDP-glucuronate; + NADH; + H |
| RP02453 | R02739 | 887 | TRIRE0005776 | Q7S986|G6PI_NEUCR | 0.0 | 5.3.1.9 | alpha-D-Glucose 6-phosphate <=> beta-D-Glucose 6-phosphate |
| RP06593 | R01528 | 709 | TRIRE0072685 | P38720|6PGD1_YEAST | 0.0 | 1.1.1.44 | NADP+; + 6-Phospho-D-gluconate <=> CO2; + NADPH; + D-Ribulose 5-phosphate + H |
| RP00216 | R01786 | 562 | TRIRE0073665 | P33284|HXK_KLULA | e-160 | 2.7.1.1 | alpha-D-Glucose + ATP; <=> alpha-D-Glucose 6-phosphate + ADP; |
Pathway diagram (see pathway with molecule structures instead)