Back to all pathways
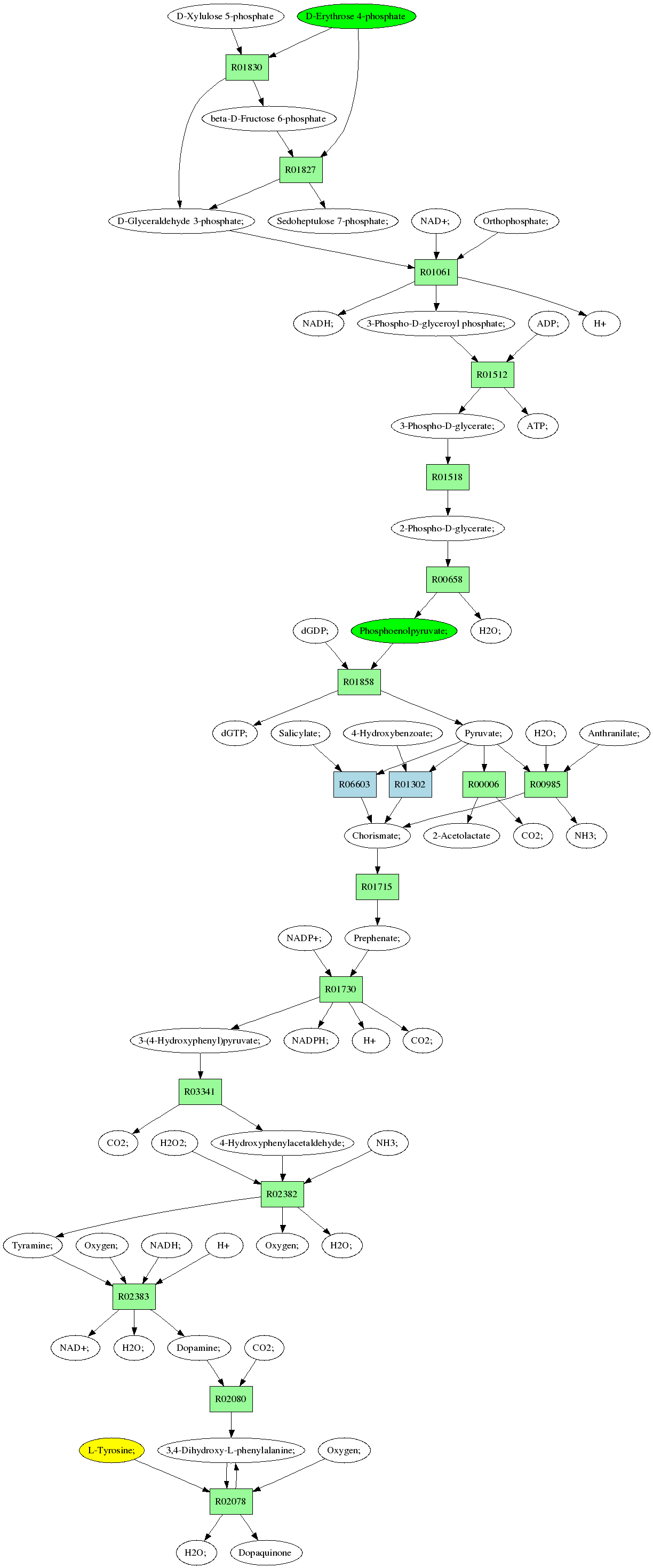
Sources: D-Erythrose 4-phosphate (C00279)
Phosphoenolpyruvate; (C00074)
Target:L-Tyrosine; (C00082)
Z: 0.333333333333
Composite map: C00074->C00082:[6->1,7->2,8->3]
Showing 1 best reactions for each rpair.
| RPAIR | Reaction | Score | Seq1 | Seq2 | Evalue | ECs | Equation |
|---|---|---|---|---|---|---|---|
| RP01568 | R01518 | 452 | TRIRE0077656 | Q747Q8|GPMI_GEOSL | e-127 | 5.4.2.1 | 2-Phospho-D-glycerate; <=> 3-Phospho-D-glycerate; |
| RP09028 | R06603 | 0 | Chorismate; <=> Salicylate; + Pyruvate; | ||||
| RP00395 | R01061 | 612 | TRIRE0119735 | P17730|G3P2_TRIKO | e-175 | 1.2.1.12 | NAD+; + D-Glyceraldehyde 3-phosphate; + Orthophosphate; <=> NADH; + 3-Phospho-D-glyceroyl phosphate; + H |
| RP13340 | R01827 | 443 | TRIRE0123026 | O42700|TAL1_SCHPO | e-124 | 2.2.1.2 | Sedoheptulose 7-phosphate; + D-Glyceraldehyde 3-phosphate; <=> D-Erythrose 4-phosphate + beta-D-Fructose 6-phosphate |
| RP01790 | R01830 | 931 | TRIRE0120635 | Q12630|TKT1_KLULA | 0.0 | 2.2.1.1 | D-Glyceraldehyde 3-phosphate; + beta-D-Fructose 6-phosphate <=> D-Xylulose 5-phosphate + D-Erythrose 4-phosphate |
| RP00113 | R01512 | 801 | TRIRE0021406 | P14228|PGK_TRIRE | 0.0 | 2.7.2.3 | 3-Phospho-D-glycerate; + ATP; <=> 3-Phospho-D-glyceroyl phosphate; + ADP; |
| RP05698 | R00006 | 793 | TRIRE0074123 | P07342|ILVB_YEAST | 0.0 | 2.2.1.6 | CO2; + 2-Acetolactate <=> Pyruvate; |
| RP01416 | R01302 | 36 | TRIRE0104059 | Q2SZY9|UBIC_BURTA | 0.095 | 4.1.3.40 | Pyruvate; + 4-Hydroxybenzoate; <=> Chorismate; |
| RP00123 | R02078 | 266 | TRIRE0050793 | P00440|TYRO_NEUCR | 7e-71 | 1.14.18.1 | Oxygen; + 3,4-Dihydroxy-L-phenylalanine; + L-Tyrosine; <=> 3,4-Dihydroxy-L-phenylalanine; + H2O; + Dopaquinone |
| RP01711 | R01715 | 234 | TRIRE0120219 | P32178|CHMU_YEAST | 2e-61 | 5.4.99.5 | Chorismate; <=> Prephenate; |
| RP06826 | R02080 | 370 | TRIRE0078552 | P20711|DDC_HUMAN | e-102 | 4.1.1.28 | 3,4-Dihydroxy-L-phenylalanine; <=> CO2; + Dopamine; |
| RP00214 | R00985 | 1023 | TRIRE0067003 | P00908|TRPG_NEUCR | 0.0 | 4.1.3.27,4.1.1.48,5.3.1.24 | Chorismate; + NH3; <=> H2O; + Anthranilate; + Pyruvate; |
| RP01215 | R00985 | 1023 | TRIRE0067003 | P00908|TRPG_NEUCR | 0.0 | 4.1.3.27,4.1.1.48,5.3.1.24 | Chorismate; + NH3; <=> H2O; + Anthranilate; + Pyruvate; |
| RP01033 | R00658 | 777 | TRIRE0120568 | Q76KF9|ENO_PENCH | 0.0 | 4.2.1.11 | 2-Phospho-D-glycerate; <=> H2O; + Phosphoenolpyruvate; |
| RP02956 | R03341 | 533 | TRIRE0121534 | P16467|PDC5_YEAST | e-151 | 4.1.1.1,4.1.1.- | 3-(4-Hydroxyphenyl)pyruvate; <=> CO2; + 4-Hydroxyphenylacetaldehyde; |
| RP01958 | R02080 | 370 | TRIRE0078552 | P20711|DDC_HUMAN | e-102 | 4.1.1.28 | 3,4-Dihydroxy-L-phenylalanine; <=> CO2; + Dopamine; |
| RP02170 | R02382 | 588 | TRIRE0056646 | P46882|AOFN_ASPNG | e-168 | 1.4.3.4 | Tyramine; + Oxygen; + H2O; <=> 4-Hydroxyphenylacetaldehyde; + NH3; + H2O2; |
| RP00334 | R01730 | 478 | TRIRE0046545 | O60078|TYR1_SCHPO | e-134 | 1.3.1.13 | NADP+; + Prephenate; <=> 3-(4-Hydroxyphenyl)pyruvate; + NADPH; + H+ + CO2; |
| RP02171 | R02383 | 413 | TRIRE0069696 | P54989|NTAA_CHEHE | e-115 | 1.14.13.- | Tyramine; + Oxygen; + NADH; + H <=> NAD+; + H2O; + Dopamine; |
| RP00036 | R01858 | 1069 | TRIRE0078439 | P31865|KPYK_TRIRE | 0.0 | 2.7.1.40 | dGTP; + Pyruvate; <=> Phosphoenolpyruvate; + dGDP; |
Pathway diagram (see pathway with molecule structures instead)