Guided tour to Pathway Assistant
This page will introduce you to the different functionalities of the Pathway Assistant portal. In the tour, you will import an example metabolic network model into the system and discover pathways between metabolites of interest. Finally, the resulting pathways will be visualised.
The following services will be discussed.
- ReMatch: integration of metabolic network models by semi-automated matching of metabolite and reaction vocabulary for the user network and the Pathway Assistant database.
-
Topology Generator:
- search for modifications that allow better target substance yield in a metabolic network
- search for simple (non-branching) pathways and feasible (branching) pathways between two metabolites.
- Metabolic Network Visualisator: visualises metabolic networks as graphs.
Getting started
Pathway Assistant is unavailable for now.
 Before using the software, you have to create a user account.
Click "Create New Account" in the upper right corner of the Pathway
Assistant front page. Please enter your information and
click the "Create New Account" button on the form to create the
account. This will take you to Pathway Assistant. One of the three
tabs, Topology Generator, is shown by default.
Before using the software, you have to create a user account.
Click "Create New Account" in the upper right corner of the Pathway
Assistant front page. Please enter your information and
click the "Create New Account" button on the form to create the
account. This will take you to Pathway Assistant. One of the three
tabs, Topology Generator, is shown by default.
Next, you can either import a metabolic network model of your own (next section) or work with one of the networks provided by the service (Topology Generator section).
ReMatch
If you wish to try out ReMatch and integrate a metabolic network of your own to the system (you can try the software with our example network too), please follow these instructions. In Pathway Assistant, ReMatch is accessed by clicking the "ReMatch" tab.
Topology Generator
Topology Generator provides computational methods for two purposes:
- Searching for modifications (insertions and deletions of reactions) in the metabolic network that provide better yield of target compounds.
- Searching for metabolic pathways from one metabolite to another. Two different kinds of pathways can be searched for. First, simple pathways are reactions in sequence that connect the two metabolites. Second, feasible pathways are self-sufficient in terms of substrates to pathway reactions (i.e., no dangling substrates).
We will cover the latter part in the following section.
Finding metabolic pathways with Topology Generator
In this section, we will search for pathways between Glucose and Pyruvate in a metabolic network. Please choose the Topology Generator tab from the top of the page, if it is not already chosen.
First, we will create a new search
task for our pathway search. Please select 'Create new search task'. A new
search task, named according to the current date and time, will appear in the
list on the left. If you wish, you can now rename the search task to
reflect the task at hand by typing the name into the Search Task Name
box and clicking 'Set search task name'.
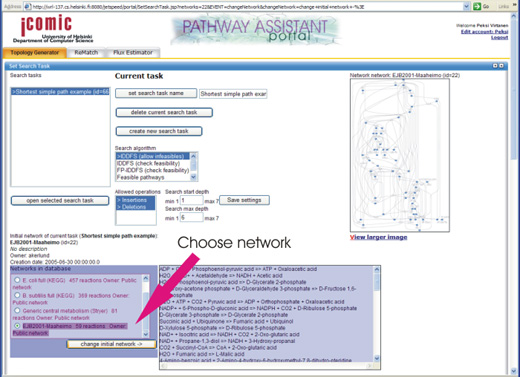
Then, we will choose a metabolic network from which pathways will
be searched for. The available networks in the database are shown in
the "Networks in database" list. If you imported a network of your own
with ReMatch, you can now choose it
from the list. Otherwise, choose the network named "EJB2001-Maaheimo".
Please choose the network from the list by clicking its radio button and click 'Change initial network'.
The box on the right should now show the reactions of this network.
Simple pathways from Glucose to Pyruvate
We will start by searching for simple pathways from Glucose to Pyruvate.
A simple pathway from metabolite A to metabolite B is just a set of reactions in sequence in such way
that metabolite A is consumed by the first reaction and metabolite B
is produced by the last reaction in the sequence.
To search for simple pathways, choose the "Shortest simple path" option from the Search Algorithm list and
click 'Save settings'.
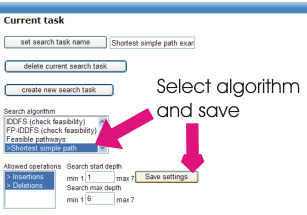
In this example, we wish only to see whether a pathway exists between Glucose and Pyruvate and not score the pathways according to the yield. Please click the 'No evaluation' button in the Search Task Parameters portlet.
The last thing to do before we can start the search is to specify Glucose and Pyruvate as the source and target metabolites, respectively. This is done in Metabolite Roles portlet. If the portlet is minimized (i.e., only its name is shown), you can open it by clicking a button on the right of the text "Metabolite Roles".
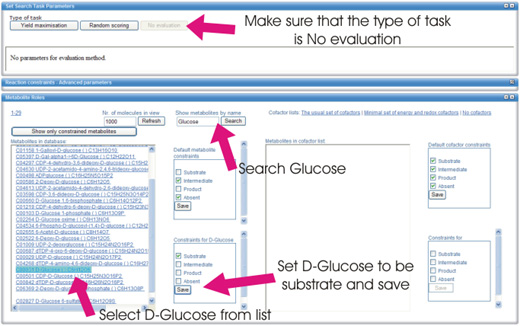 In the "Show metabolites by name" box, enter 'Glucose' and click
"Search" button. From the list of metabolites, click
"D-Glucose". You can now specify Glucose as the source by selecting
"Substrate" in the "Constraints for D-Glucose" box and clicking
"Save".
In similar manner, set a Product constraint to 'Pyruvic acid'.
In the "Show metabolites by name" box, enter 'Glucose' and click
"Search" button. From the list of metabolites, click
"D-Glucose". You can now specify Glucose as the source by selecting
"Substrate" in the "Constraints for D-Glucose" box and clicking
"Save".
In similar manner, set a Product constraint to 'Pyruvic acid'.
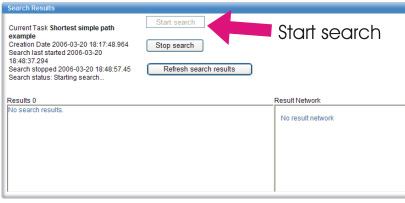 We can now start the search. In the bottom Search Results portlet, click "Start search" button. After a few seconds,
please click the "Refresh search results" button to display the results.
The result pathways are shown in the left panel. You should note that
the first result is always the metabolic network chosen in the Search
Task portlet. If a pathway was found, it is the second result.
We can now start the search. In the bottom Search Results portlet, click "Start search" button. After a few seconds,
please click the "Refresh search results" button to display the results.
The result pathways are shown in the left panel. You should note that
the first result is always the metabolic network chosen in the Search
Task portlet. If a pathway was found, it is the second result.
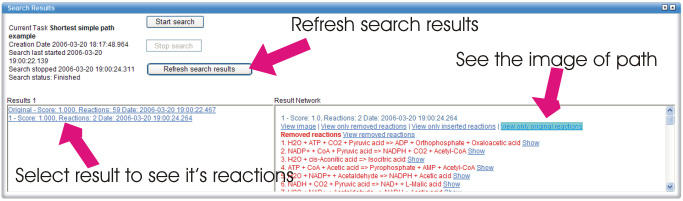
You can now click on the second result to view the pathway. A list of
pathway reactions is displayed on the right panel.
The list of reactions on the pathway is separated in three parts:
Removed, inserted and original reactions. These parts show the
reactions in relation to the initial network. For instance, the list
of removed reactions shows reactions that were in the initial network
but are not on the resulting pathway. In our example, we are
interested in reactions in the original reactions list: these are the
reactions on the result pathway. Scroll past the removed reaction part
to see the reactions in the result.
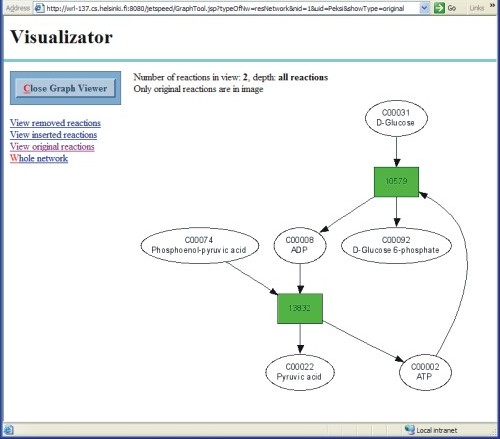
If you selected "EJB2001-Maaheimo" as the initial network, you should now have the following pathway of two reactions:
1. ATP + D-Glucose => ADP + D-Glucose 6-phosphate 2. ADP + Phosphoenol-pyruvic acid => ATP + Pyruvic acid
You can view the pathway by clicking "View original reactions". We will discuss the options for visualisation of results in a separate section.
In the previous example, Glucose and Pyruvate were connected via ADP. Highly connected metabolites, such as energy and redox cofactors, usually participate on the shortest paths between any two metabolites. To avoid this, you can choose a list of metabolites to avoid in the path finding. The list is chosen in Metabolite Roles portlet by clicking on some of the Cofactor lists in the right hand side of the portlet. For example, try "The usual set of cofactors". The metabolites on the list should be displayed in the box below.
Now, you can restart the search by clicking "Start search" in the "Search Results" portlet. After a few seconds, click "Refresh search results" to view the new results.
Feasible pathways from Glucose to Pyruvate
A feasible pathway from metabolite A to metabolite B is a set of reactions in which all reaction substrates, excluding metabolite A, are by-products of the reactions on the pathway. In other words, a feasible pathway is able to produce any metabolite required by any reaction on the pathway.
In addition to simple paths, you can also search for feasible pathways by selecting "Feasible pathways" as the search algorithm.
For detailed discussion on feasible pathways, please see
Esa Pitkänen, Ari Rantanen, Juho Rousu and Esko Ukkonen:
Finding Feasible Pathways in Metabolic Networks.
10th Panhellenic Conference on Informatics 2005, Volos, Greece.
Metabolic Network Visualisator
Metabolic Network Visualisator allows you to view a graphical representation of metabolic networks in the system. To view a network in the database, click on the "View larger image" link below the thumbnail image of the currently selected initial network in "Search Task" portlet. To view a result network from a search, click on "View image" link in the Result Network panel in the Search Result portlet. By clicking on "View only removed reactions", "View only inserted reactions" or "View only original reactions", you can view only reactions that are not in the initial network, missing from the initial network or the same as in the initial network, respectively.
In the graph view, reactions are drawn as boxes and metabolites as ellipses. In particular, removed reactions are shown in red, inserted reactions in purple and original reactions in green. You can choose to view only removed, inserted or original reactions at a time by clicking on appropriate links in the left panel.
You can display information on an individual metabolite or reaction by clicking it on the graph. For metabolites, the name and link to the KEGG database is shown on the left. In the top panel, the structure of the metabolite is shown. For reactions, structures of all the substrates and products of the reaction are shown in the top panel.
Previous update: 19.10.2009 23:14 - Esa Pitkänen

