Biomine search engine prototype
Try out the search engine at biomine.cs.helsinki.fi https://biomine.ijs.si.
More information + citation:
Biomine: Predicting links between biological entities using network models of heterogeneous database. Lauri MA Eronen and Hannu TT Toivonen. BMC Bioinformatics 13:119, 2012.
Biomine project: Knowledge discovery in biological databases
Public biological databases contain huge amounts of rich data, such as annotated sequences, proteins, domains, and orthology groups, genes and gene expressions, gene and protein interactions, scientific articles, and ontologies. The Biomine project develops methods for the analysis of such collections of data.
Example problem: candidate gene analysis
As a motivating problem, consider gene mapping. Mapping of a disease can result in tens or hundreds of candidate genes. The next problem is then to identify the most promising genes for further research. The current state of the art consists largely of manual exploration of public databases, for instance to find connections between genes and phenotypes. The Biomine project develops methods for automated discovery and prediction of previously unknown and potentially biologically relevant connections. Our focus is on candidate gene analysis, and methods we develop help geneticists assess the potential relationship of their candidate genes to the disease under study.
Research approach: graph mining
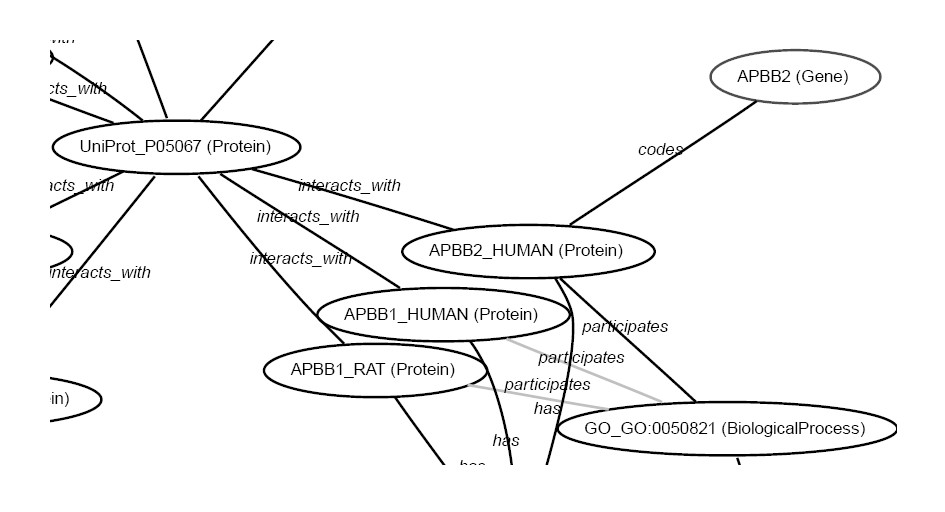
In the Biomine approach, all information is handled as graphs: nodes correspond to different concepts (such as gene, protein, domain, phenotype, biological process, tissue), and semantically labelled edges connect related concepts (e.g., gene BCHE codes protein CHLE, which in turn has the molecular function 'beta-amyloid binding'; see a simple example graph). One central goal is to develop methods for establishing new, previously unknown connections between nodes, in other words, creation of biological hypotheses. We develop and use data mining algorithms for this. Predicted connections could be based, for instance, on discovered analogies between two concepts or their contexts, or on finding (strong) paths between concepts.
Applications for graph mining
Discovery of patterns in graphs have numerous potential applications in biology, including the analysis of metabolic networks, regulatory relationships, protein structures, and chemical compounds, as obvious candidates. Virtually any data could be described as graphs, and the developed methods can potentially be applied in other areas, too.
People and partners
Researchers in the project:
- Prof. Hannu Toivonen (contact person),
- Lauri Eronen,
- Petteri Hintsanen,
- Kimmo Kulovesi, and
- Laura Langohr.
The project was carried out by the Discovery Group. in the HIIT Basic Research Unit at the Department of Computer Science, University of Helsinki. The project has been funded by
- Tekes, the Finnish Funding Agency for Technology and Innovation,
- Jurilab Ltd,
- Biocomputing Platforms Ltd, and
- GeneOS Ltd,
and it co-operated with
- Prof. Juha Kere, Department of Medical Genetics, University of Helsinki and Karolinska Institutet, Stockholm
- CSC, the Finnish IT center for science and
- VTT Biotechnology.
Publications
-
Biomine: Predicting links between biological entities
using network models of heterogeneous database
Lauri Eronen and Hannu Toivonen. BMC Bioinformatics 13:119, 2012. -
SegMine workflows for semantic microarray data analysis in Orange4WS
Vid Podpecan, Nada Lavrac, Igor Mozetic, Petra Kralj Novak, Igor Trajkovski, Laura Langohr, Kimmo Kulovesi, Hannu Toivonen, Marko Petek, Helena Motaln, Kristina Gruden. BMC Bioinformatics 12:416, 2011. -
Fast Discovery of Reliable Subnetworks
Petteri Hintsanen, Hannu Toivonen, and Petteri Sevon. The 2010 International Conference on Advances in Social Networks Analysis and Mining (ASONAM), 104-111, Odense, Denmark, August 2010. -
Fast Discovery of Reliable k-terminal Subgraphs
Melissa Kasari, Hannu Toivonen, and Petteri Hintsanen. The 14th Pacific-Asia Conference on Knowledge Discovery and Data Mining (PAKDD), Part II, LNAI 6119, 168-177, Hyderabad, India, June 2010. -
Bisociative Knowledge Discovery for Microarray Data Analysis
Igor Mozetic, Nada Lavrac, Vid Podpecan, Petra Kralj Novak, Helena Motaln, Marko Petek, Kristina Gruden, Hannu Toivonen, and Kimmo Kulovesi. The 1st International Conference on Computational Creativity (ICCC-X), 190-199, Lisbon, Portugal, January 2010. -
Finding Reliable Subgraphs from Large Probabilistic Graphs
Petteri Hintsanen, Hannu Toivonen. Data Mining and Knowledge Discovery 17 (1): 3-23. 2008. -
Compressing Probabilistic Prolog Programs
Luc De Raedt, Kristian Kersting, Angelika Kimmig, Kate Revoredo, Hannu Toivonen. Machine Learning 70 (2-3): 151-168. 2008. -
Subgraph Queries by Context-free Grammars
Petteri Sevon and Lauri Eronen. Journal of Integrative Bioinformatics 5 (2): 100. 2008. - Probabilistic Explanation Based Learning
Luc De Raedt, Angelika Kimmig, Hannu Toivonen. 18th European Conference on Machine Learning (ECML), 176-187, Warsaw, Poland, September 2007. Winner of the ECML-07 Best Paper Award. - The Most Reliable Subgraph Problem, Petteri Hintsanen. 11th European Conference on Principles and Practice of Knowledge Discovery in Databases (PKDD), 471-478, Warsaw, Poland, September 2007.
-
ProbLog: A Probabilistic Prolog and its Application in Link Discovery
Luc De Raedt, Angelika Kimmig, Hannu Toivonen. Twentieth International Joint Conference on Artificial Intelligence (IJCAI-07), 2468-2473, Hyderabad, India, January 2007. -
Link discovery in graphs derived from biological databases
Petteri Sevon, Lauri Eronen, Petteri Hintsanen, Kimmo Kulovesi, Hannu Toivonen. 3rd International Workshop on Data Integration in the Life Sciences 2006 (DILS'06), LNBI 4705, 35-49, Hinxton, UK, July 2006. Springer.
Research Group
The project is carried by the Discovery Research Group.
Contact
Contact: Prof. Hannu Toivonen, email firstname.lastname@cs.helsinki.fi.
Up to: Department of Computer Science | HIIT